Active Learning
Active Learning
Active learning is a special case of machine learning in which a learning algorithm is able to interactively query the user (or some other information source called the oracle to obtain the desired outputs at new data points. There are situations in which unlabeled data is abundant but manually labeling is expensive. In such a scenario, learning algorithms can actively query the user/teacher for labels. This type of iterative supervised learning is called active learning. Since the learner chooses the examples, the number of examples to learn a concept can often be much lower than the number required in normal supervised learning.
There are three scenarios for Active Learning:
- Membership query synthesis, i.e., a generated sample is sent to an oracle for labeling.
- Stream-Based selective sampling, i.e, each sample is considered separately -in our case for label-querying or rejection. Similarly to online-learning, the data is not saved, there are no assumptions on data distribution, and therefore it is adaptive to change.
- Pool-Based sampling, i.e., sampled are chosen from a pool of unlabeled data for the purpose of labeling.


Uncertainty sampling
All active learning scenarios involve evaluating the informativeness of unlabeled instances, which can either be generated de novo or sampled from a given distribution.
Perhaps the simplest and most commonly used query framework is uncertainty sampling(Lewis and Gale, 1994). In this framework, an active learner queries the instances about which it is least certain how to label. This approach is often straightforward for probabilistic learning models.
- Least Confidence (LC): in this strategy, the learner selects the instance for which it has the least confidence in its most likely label.
- Margin Sampling: the shortcoming of the LC strategy, is that it only takes into consideration the most probable label and disregards the other label probabilities. The margin sampling strategy seeks to overcome this disadvantage by selecting the instance that has the smallest difference between the first and second most probable labels.
- Entropy Sampling: in order to utilize all the possible label probabilities, you use a popular measure called entropy. The entropy formula is applied to each instance and the instance with the largest value is queried.
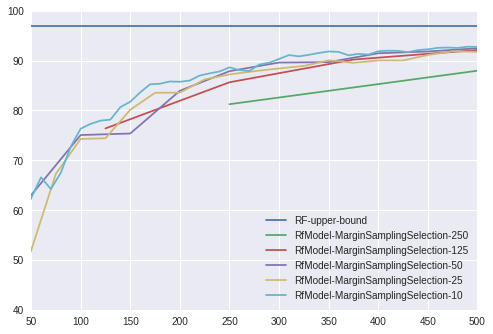
We compare several learning algorithms, such as support vector machine (SVM) with a linear kernel, random forest (RF) and logistic regression (LOG). Each algorithm was executed with all of the selection functions using all ‘k’ = [10,25,50,125,250], accumulating a total of 80 experiment on MNIST. For this example the random forest classifier with hyperparameter k=10 performs best.
Reference
- Olsson, Fredrik (April 2009). “A literature survey of active machine learning in the context of natural language processing”. SICS Technical Report T2009:06.
- Settles, Burr (2010). “Active Learning Literature Survey” (PDF). Computer Sciences Technical Report 1648. University of Wisconsin–Madison. Retrieved 2014-11-18.
import os
import time
import json
import pickle
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.mlab as mlab
from scipy import stats
from pylab import rcParams
from sklearn.utils import check_random_state
from sklearn.datasets import load_digits
from sklearn.datasets import fetch_mldata
from sklearn.decomposition import PCA
from sklearn.preprocessing import scale
from sklearn.preprocessing import MinMaxScaler
from sklearn.cluster import KMeans
from sklearn.mixture import GaussianMixture
from sklearn.svm import LinearSVC, SVC
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import LogisticRegression
from sklearn import metrics
from sklearn.metrics import pairwise_distances_argmin_min
from sklearn.metrics import confusion_matrix
from sklearn.metrics import precision_recall_curve
from sklearn.metrics import average_precision_score
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier, \
GradientBoostingClassifier
trainset_size = 60000 # ie., testset_size = 10000
max_queried = 500
# ==============================================================================
def download():
mnist = fetch_mldata('MNIST original')
X = mnist.data.astype('float64')
y = mnist.target
print ('MNIST:', X.shape, y.shape)
return (X, y)
def split(train_size):
X_train_full = X[:train_size]
y_train_full = y[:train_size]
X_test = X[train_size:]
y_test = y[train_size:]
return (X_train_full, y_train_full, X_test, y_test)
# ==============================================================================
class BaseModel(object):
def __init__(self):
pass
def fit_predict(self):
pass
class SvmModel(BaseModel):
model_type = 'Support Vector Machine with linear Kernel'
def fit_predict(self, X_train, y_train, X_val, X_test, c_weight):
print ('training svm...')
self.classifier = SVC(C=1, kernel='linear', probability=True,
class_weight=c_weight)
self.classifier.fit(X_train, y_train)
self.test_y_predicted = self.classifier.predict(X_test)
self.val_y_predicted = self.classifier.predict(X_val)
return (X_train, X_val, X_test, self.val_y_predicted,
self.test_y_predicted)
class GmmModel(BaseModel):
model_type = 'Gaussian Mixture Model'
def fit_predict(self, X_train, y_train, X_val, X_test, c_weight):
print ('training gaussian mixture model...')
pca = PCA(n_components=75).fit(X_train) # ,whiten=True).fit(X_train)
reduced_train_data = pca.transform(X_train)
reduced_test_data = pca.transform(X_test)
reduced_val_data = pca.transform(X_val)
print ('PCA: explained_variance_ratio_',
np.sum(pca.explained_variance_ratio_))
self.classifier = GaussianMixture(n_components=10, covariance_type='full')
self.classifier.fit(reduced_train_data)
self.test_y_predicted = \
self.classifier.predict(reduced_test_data)
self.val_y_predicted = self.classifier.predict(reduced_val_data)
return (reduced_train_data, reduced_val_data,
reduced_test_data, self.val_y_predicted,
self.test_y_predicted)
class LogModel(BaseModel):
model_type = 'Multinominal Logistic Regression'
def fit_predict(self, X_train, y_train, X_val, X_test, c_weight):
print ('training multinomial logistic regression')
train_samples = X_train.shape[0]
self.classifier = LogisticRegression(
C=50. / train_samples,
multi_class='multinomial',
penalty='l1',
solver='saga',
tol=0.1,
class_weight=c_weight,
)
self.classifier.fit(X_train, y_train)
self.test_y_predicted = self.classifier.predict(X_test)
self.val_y_predicted = self.classifier.predict(X_val)
return (X_train, X_val, X_test, self.val_y_predicted,
self.test_y_predicted)
class GbcModel(BaseModel):
model_type = 'Gradient Boosting Classifier'
def fit_predict(self, X_train, y_train, X_val, X_test, c_weight):
print ('training gradient boosting...')
parm = {
'n_estimators': 1200,
'max_depth': 3,
'subsample': 0.5,
'learning_rate': 0.01,
'min_samples_leaf': 1,
'random_state': 3,
}
self.classifier = GradientBoostingClassifier(**parm)
self.classifier.fit(X_train, y_train)
self.test_y_predicted = self.classifier.predict(X_test)
self.val_y_predicted = self.classifier.predict(X_val)
return (X_train, X_val, X_test, self.val_y_predicted,
self.test_y_predicted)
class RfModel(BaseModel):
model_type = 'Random Forest'
def fit_predict(self, X_train, y_train, X_val, X_test, c_weight):
print ('training random forest...')
self.classifier = RandomForestClassifier(n_estimators=500, class_weight=c_weight)
self.classifier.fit(X_train, y_train)
self.test_y_predicted = self.classifier.predict(X_test)
self.val_y_predicted = self.classifier.predict(X_val)
return (X_train, X_val, X_test, self.val_y_predicted, self.test_y_predicted)
# ====================================================================================================
class TrainModel:
def __init__(self, model_object):
self.accuracies = []
self.model_object = model_object()
def print_model_type(self):
print (self.model_object.model_type)
# we train normally and get probabilities for the validation set. i.e., we use the probabilities to select the most uncertain samples
def train(self, X_train, y_train, X_val, X_test, c_weight):
print ('Train set:', X_train.shape, 'y:', y_train.shape)
print ('Val set:', X_val.shape)
print ('Test set:', X_test.shape)
t0 = time.time()
(X_train, X_val, X_test, self.val_y_predicted,
self.test_y_predicted) = \
self.model_object.fit_predict(X_train, y_train, X_val, X_test, c_weight)
self.run_time = time.time() - t0
return (X_train, X_val, X_test) # we return them in case we use PCA, with all the other algorithms, this is not needed.
# we want accuracy only for the test set
def get_test_accuracy(self, i, y_test):
classif_rate = np.mean(self.test_y_predicted.ravel() == y_test.ravel()) * 100
self.accuracies.append(classif_rate)
print('--------------------------------')
print('Iteration:',i)
print('--------------------------------')
print('y-test set:',y_test.shape)
print('Example run in %.3f s' % self.run_time,'\n')
print("Accuracy rate for %f " % (classif_rate))
print("Classification report for classifier %s:\n%s\n" % (self.model_object.classifier, metrics.classification_report(y_test, self.test_y_predicted)))
print("Confusion matrix:\n%s" % metrics.confusion_matrix(y_test, self.test_y_predicted))
print('--------------------------------')
# ====================================================================================================
def get_k_random_samples(initial_labeled_samples, X_train_full,
y_train_full):
random_state = check_random_state(0)
permutation = np.random.choice(trainset_size,
initial_labeled_samples,
replace=False)
print ()
print ('initial random chosen samples', permutation.shape),
# permutation)
X_train = X_train_full[permutation]
y_train = y_train_full[permutation]
X_train = X_train.reshape((X_train.shape[0], -1))
bin_count = np.bincount(y_train.astype('int64'))
unique = np.unique(y_train.astype('int64'))
print (
'initial train set:',
X_train.shape,
y_train.shape,
'unique(labels):',
bin_count,
unique,
)
return (permutation, X_train, y_train)
# ====================================================================================================
class BaseSelectionFunction(object):
def __init__(self):
pass
def select(self):
pass
class RandomSelection(BaseSelectionFunction):
@staticmethod
def select(probas_val, initial_labeled_samples):
random_state = check_random_state(0)
selection = np.random.choice(probas_val.shape[0], initial_labeled_samples, replace=False)
# print('uniques chosen:',np.unique(selection).shape[0],'<= should be equal to:',initial_labeled_samples)
return selection
class MinStdSelection(BaseSelectionFunction):
# select the samples where the std is smallest - i.e., there is uncertainty regarding the relevant class
# and then train on these "hard" to classify samples.
@staticmethod
def select(probas_val, initial_labeled_samples):
std = np.std(probas_val * 100, axis=1)
selection = std.argsort()[:initial_labeled_samples]
selection = selection.astype('int64')
# print('std',std.shape,std)
# print()
# print('selection',selection, selection.shape, std[selection])
return selection
class MarginSamplingSelection(BaseSelectionFunction):
@staticmethod
def select(probas_val, initial_labeled_samples):
rev = np.sort(probas_val, axis=1)[:, ::-1]
values = rev[:, 0] - rev[:, 1]
selection = np.argsort(values)[:initial_labeled_samples]
return selection
class EntropySelection(BaseSelectionFunction):
@staticmethod
def select(probas_val, initial_labeled_samples):
e = (-probas_val * np.log2(probas_val)).sum(axis=1)
selection = (np.argsort(e)[::-1])[:initial_labeled_samples]
return selection
# ====================================================================================================
class Normalize(object):
def normalize(self, X_train, X_val, X_test):
self.scaler = MinMaxScaler()
X_train = self.scaler.fit_transform(X_train)
X_val = self.scaler.transform(X_val)
X_test = self.scaler.transform(X_test)
return (X_train, X_val, X_test)
def inverse(self, X_train, X_val, X_test):
X_train = self.scaler.inverse_transform(X_train)
X_val = self.scaler.inverse_transform(X_val)
X_test = self.scaler.inverse_transform(X_test)
return (X_train, X_val, X_test)
# ====================================================================================================
class TheAlgorithm(object):
accuracies = []
def __init__(self, initial_labeled_samples, model_object, selection_function):
self.initial_labeled_samples = initial_labeled_samples
self.model_object = model_object
self.sample_selection_function = selection_function
def run(self, X_train_full, y_train_full, X_test, y_test):
# initialize process by applying base learner to labeled training data set to obtain Classifier
(permutation, X_train, y_train) = \
get_k_random_samples(self.initial_labeled_samples,
X_train_full, y_train_full)
self.queried = self.initial_labeled_samples
self.samplecount = [self.initial_labeled_samples]
# permutation, X_train, y_train = get_equally_k_random_samples(self.initial_labeled_samples,classes)
# assign the val set the rest of the 'unlabelled' training data
X_val = np.array([])
y_val = np.array([])
X_val = np.copy(X_train_full)
X_val = np.delete(X_val, permutation, axis=0)
y_val = np.copy(y_train_full)
y_val = np.delete(y_val, permutation, axis=0)
print ('val set:', X_val.shape, y_val.shape, permutation.shape)
print ()
# normalize data
normalizer = Normalize()
X_train, X_val, X_test = normalizer.normalize(X_train, X_val, X_test)
self.clf_model = TrainModel(self.model_object)
(X_train, X_val, X_test) = self.clf_model.train(X_train, y_train, X_val, X_test, 'balanced')
active_iteration = 1
self.clf_model.get_test_accuracy(1, y_test)
# fpfn = self.clf_model.test_y_predicted.ravel() != y_val.ravel()
# print(fpfn)
# self.fpfncount = []
# self.fpfncount.append(fpfn.sum() / y_test.shape[0] * 100)
while self.queried < max_queried:
active_iteration += 1
# get validation probabilities
probas_val = \
self.clf_model.model_object.classifier.predict_proba(X_val)
print ('val predicted:',
self.clf_model.val_y_predicted.shape,
self.clf_model.val_y_predicted)
print ('probabilities:', probas_val.shape, '\n',
np.argmax(probas_val, axis=1))
# select samples using a selection function
uncertain_samples = \
self.sample_selection_function.select(probas_val, self.initial_labeled_samples)
# normalization needs to be inversed and recalculated based on the new train and test set.
X_train, X_val, X_test = normalizer.inverse(X_train, X_val, X_test)
# get the uncertain samples from the validation set
print ('trainset before', X_train.shape, y_train.shape)
X_train = np.concatenate((X_train, X_val[uncertain_samples]))
y_train = np.concatenate((y_train, y_val[uncertain_samples]))
print ('trainset after', X_train.shape, y_train.shape)
self.samplecount.append(X_train.shape[0])
bin_count = np.bincount(y_train.astype('int64'))
unique = np.unique(y_train.astype('int64'))
print (
'updated train set:',
X_train.shape,
y_train.shape,
'unique(labels):',
bin_count,
unique,
)
X_val = np.delete(X_val, uncertain_samples, axis=0)
y_val = np.delete(y_val, uncertain_samples, axis=0)
print ('val set:', X_val.shape, y_val.shape)
print ()
# normalize again after creating the 'new' train/test sets
normalizer = Normalize()
X_train, X_val, X_test = normalizer.normalize(X_train, X_val, X_test)
self.queried += self.initial_labeled_samples
(X_train, X_val, X_test) = self.clf_model.train(X_train, y_train, X_val, X_test, 'balanced')
self.clf_model.get_test_accuracy(active_iteration, y_test)
print ('final active learning accuracies',
self.clf_model.accuracies)
# get MNIST
(X, y) = download()
(X_train_full, y_train_full, X_test, y_test) = split(trainset_size)
print ('train:', X_train_full.shape, y_train_full.shape)
print ('test :', X_test.shape, y_test.shape)
classes = len(np.unique(y))
print ('unique classes', classes)
def pickle_save(fname, data):
filehandler = open(fname,"wb")
pickle.dump(data,filehandler)
filehandler.close()
print('saved', fname, os.getcwd(), os.listdir())
def experiment(d, models, selection_functions, Ks, repeats, contfrom):
algos_temp = []
print ('stopping at:', max_queried)
count = 0
for model_object in models:
if model_object.__name__ not in d:
d[model_object.__name__] = {}
for selection_function in selection_functions:
if selection_function.__name__ not in d[model_object.__name__]:
d[model_object.__name__][selection_function.__name__] = {}
for k in Ks:
d[model_object.__name__][selection_function.__name__][k] = []
for i in range(0, repeats):
count+=1
if count >= contfrom:
print ('Count = %s, using model = %s, selection_function = %s, k = %s, iteration = %s.' % (count, model_object.__name__, selection_function.__name__, k, i))
alg = TheAlgorithm(k,
model_object,
selection_function
)
alg.run(X_train_full, y_train_full, X_test, y_test)
d[model_object.__name__][selection_function.__name__][k].append(alg.clf_model.accuracies)
fname = 'Active-learning-experiment-' + str(count) + '.pkl'
pickle_save(fname, d)
if count % 5 == 0:
print(json.dumps(d, indent=2, sort_keys=True))
print ()
print ('---------------------------- FINISHED ---------------------------')
print ()
return d
max_queried = 500
# max_queried = 20
repeats = 1
models = [SvmModel, RfModel, LogModel]#, GbcModel]
# models = [RfModel, SvmModel]
selection_functions = [RandomSelection, MarginSamplingSelection, EntropySelection]#, MinStdSelection]
# selection_functions = [MarginSamplingSelection]
Ks = [250,125,50,25,10]
# Ks = [10]
d = {}
stopped_at = -1
# stopped_at = 73
# d = pickle_load('Active-learning-experiment-'+ str(stopped_at) +'.pkl')
# print(json.dumps(d, indent=2, sort_keys=True))
d = experiment(d, models, selection_functions, Ks, repeats, stopped_at+1)
print(json.dumps(d, indent=2, sort_keys=True))